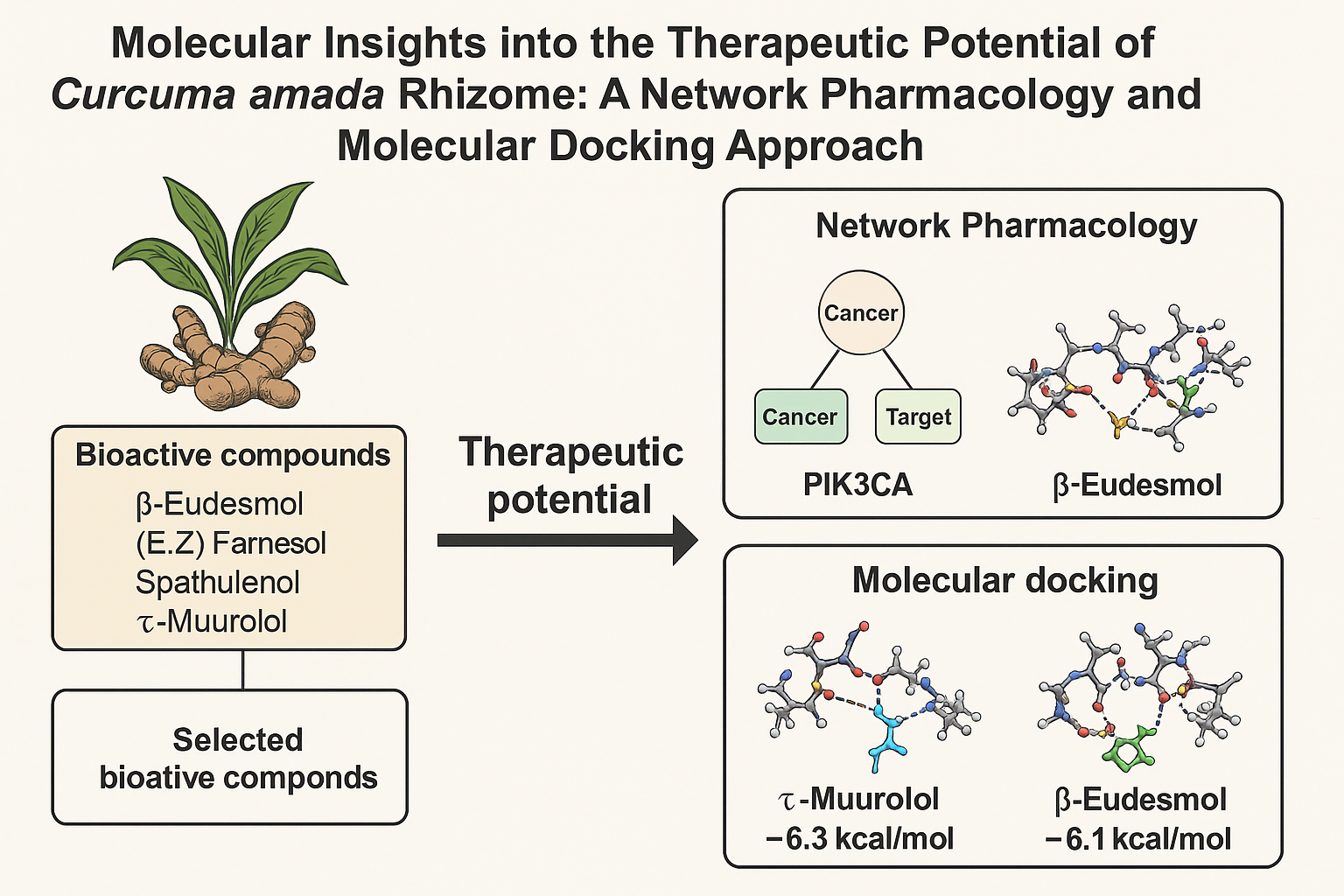
Molecular Insights into the Therapeutic Potential of Curcuma amada Rhizome: A Network Pharmacology and Docking Approach
Abstract
The rhizome of Curcuma amada (ginger mango) has traditionally been used for various medicinal purposes. However, its molecular targets and mechanisms of action are still poorly understood. This study aimed to predict the potential bioactive compounds and target proteins of C. amada using network pharmacology and molecular docking approaches. A total of 110 compounds were identified, and their predicted targets were analyzed through a protein-protein interaction (PPI) network, enrichment, and disease analysis. Key targets include phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (PIK3CA) and tyrosine-protein kinase JAK2 (JAK2), both of which are closely related to cancer-related pathways. Four compounds, β-eudesmol, (E,Z)-farnesol, spathulenol, and τ-muurolol, were selected for molecular docking studies. Validation of the docking protocol through re-docking showed low RMSD values (0.667 Å for PIK3CA and 0.474 Å for JAK2), confirming the reliability of the method. The docking results demonstrated that the native ligands and selected compounds formed multiple hydrogen bonds and extensive hydrophobic interactions with key residues in the active site. Notably, most of the interactions were hydrophobic, which is consistent with the volatile nature of the ligands. The binding affinity was below –5 kcal/mol for all tested compounds. These findings suggest that C. amada rhizomes contain bioactive compounds capable of modulating cancer-related targets, thus providing a molecular basis for their potential therapeutic effects.
Full text article
References
Ardito, F., Giuliani, M., Perrone, D., Troiano, G., & Muzio, L. Lo. (2017). The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). International Journal of Molecular Medicine, 40(2), 271. https://doi.org/10.3892/IJMM.2017.3036
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The Protein Data Bank. In Nucleic Acids Research (Vol. 28, Issue 1, pp. 235–242). https://doi.org/10.1093/nar/28.1.235
Beserra Santos, L. K., Alves Veras, M. D., Gadelha Marques, K. K., de Moraes Alves, M. M., Nogueira Mendes, A., de Amorim Carvalho, F. A., Vieira Sobral, M., Helena Chaves, M., & Ramos Gonçalves, J. C. (2020). Assessment of In Vitro Anti-melanoma Potential of Ephedranthus pisocarpus R.E.Fr. Anticancer Research, 40(9), 5015–5024. https://doi.org/10.21873/ANTICANRES.14504
BIOVIA Dassault Systèmes. (2021). BIOVIA Discovery Studio. https://www.3ds.com/biovia
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., & Lin, C. Y. (2014). cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Systems Biology, 8(4), 1–7. https://doi.org/10.1186/1752-0509-8-S4-S11/TABLES/4
Choudhary, L., Jain, R., & Sahu, S. (2024). Mango Ginger - Curcuma amada - The Uncommon Spice with Uncommon Pharmacotherapeutic Potentials. In Research Journal of Pharmacy and Technology (Vol. 17, Issue 3, pp. 1418–1424). Journal of Ravishankar University (Part-B). https://doi.org/10.52711/0974-360X.2024.00225
Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7(January), 1–13. https://doi.org/10.1038/srep42717
Daina, A., Michielin, O., & Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Research, 47(W1), W357–W3664. https://doi.org/10.1093/nar/gkz382
Hanwell, M. D., Curtis, D. E., Lonie, D. C., Vandermeerschd, T., Zurek, E., & Hutchison, G. R. (2012). Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics, 4(8), 1–17. https://doi.org/10.1186/1758-2946-4-17
He, Y., Sun, M. M., Zhang, G. G., Yang, J., Chen, K. S., Xu, W. W., & Li, B. (2021). Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduction and Targeted Therapy 2021 6:1, 6(1), 1–17. https://doi.org/10.1038/s41392-021-00828-5
Hopkins, A. L. (2008). Network pharmacology: The next paradigm in drug discovery. In Nature Chemical Biology (Vol. 4, Issue 11, pp. 682–690). Nature Publishing Group. https://doi.org/10.1038/nchembio.118
Hu, X., li, J., Fu, M., Zhao, X., & Wang, W. (2021). The JAK/STAT signaling pathway: from bench to clinic. Signal Transduction and Targeted Therapy 2021 6:1, 6(1), 1–33. https://doi.org/10.1038/s41392-021-00791-1
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., & Bolton, E. E. (2022). PubChem 2023 update. Nucleic Acids Research, 51(D1). https://doi.org/10.1093/nar/gkac956
Kumar, D., Joshi, M., Goyal, P. K., & Jayabalan, G. (2024). History and A Phytopharmacological Review of Curcuma amada. International Journal For Multidisciplinary Research, 6(1). https://doi.org/10.36948/ijfmr.2024.v06i01.12914
Lee, J. H., Chinnathambi, A., Alharbi, S. A., Shair, O. H. M., Sethi, G., & Ahn, K. S. (2019). Farnesol abrogates epithelial to mesenchymal transition process through regulating Akt/mTOR pathway. Pharmacological Research, 150, 104504. https://doi.org/10.1016/J.PHRS.2019.104504
Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today: Technologies, 1(4), 337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Lipinski, C. A. (2016). Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. In Advanced Drug Delivery Reviews (Vol. 101, pp. 34–41). Adv Drug Deliv Rev. https://doi.org/10.1016/j.addr.2016.04.029
Mahadevi, R., & Kavitha, R. (2020). Phytochemical and pharmacological properties of Curcuma amada: A review. In International Journal of Research in Pharmaceutical Sciences (Vol. 11, Issue 3, pp. 3546–3555). J. K. Welfare and Pharmascope Foundation. https://doi.org/10.26452/ijrps.v11i3.2510
Manjima, R. B., Ramya, S., Kavithaa, K., Paulpandi, M., Saranya, T., Harysh Winster, S. B., Balachandar, V., & Arul, N. (2021). Spathulenol attenuates 6-hydroxydopamine induced neurotoxicity in SH-SY5Y neuroblastoma cells. Gene Reports, 25, 101396. https://doi.org/10.1016/J.GENREP.2021.101396
Meidatuzzahra, D., & Swandayani, R. E. (2020). Pemanfaatan Famili Zingiberaceae Sebagai Obat Tradisional oleh Masyarakat Suku Sasak di Desa Suranadi, Kecamatan Narmada, Kabupaten Lombok Barat. Bionature, 21(2). https://doi.org/10.35580/bionature.v21i2.16494
Morris, G. M., Ruth, H., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. https://doi.org/10.1002/jcc.21256
Nepusz, T., Yu, H., & Paccanaro, A. (2012). Detecting overlapping protein complexes in protein-protein interaction networks. Nature Methods, 9(5), 471. https://doi.org/10.1038/NMETH.1938
Nurainy, F., Utomo, T. P., Susilowati, & Meindari, L. (2019). Karakteristik Kimia dan Sensori Permen Jelly Temu Mangga (Curcuma mangga Val.) pada Berbagai Proporsi Penambahan Sari Buah Mangga Kuweni (Mangifera odorata Griff). Semiloka Nasional FKPTPI, Universitas Padjajaran, 20, 372384.
Owen, K. L., Brockwell, N. K., & Parker, B. S. (2019). JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers 2019, Vol. 11, Page 2002, 11(12), 2002. https://doi.org/10.3390/CANCERS11122002
Padalia, R. C., Verma, R. S., Sundaresan, V., Chauhan, A., Chanotiya, C. S., & Yadav, A. (2013). Volatile terpenoid compositions of leaf and rhizome of Curcuma amada Roxb. from Northern India. Journal of Essential Oil Research, 25(1), 17–22. https://doi.org/10.1080/10412905.2012.747271;PAGE:STRING:ARTICLE/CHAPTER
Park, J. S., Kwon, J. K., Kim, H. R., Kim, H. J., Kim, B. S., & Jung, J. Y. (2014). Farnesol induces apoptosis of DU145 prostate cancer cells through the PI3K/Akt and MAPK pathways. International Journal of Molecular Medicine, 33(5), 1169–1176. https://doi.org/10.3892/ijmm.2014.1679
Policegoudra, R. S., Rehna, K., Rao, L. J., & Aradhya, S. M. (2010). Antimicrobial, antioxidant, cytotoxicity and platelet aggregation inhibitory activity of a novel molecule isolated and characterized from mango ginger (Curcuma amada Roxb.) rhizome. Journal of Biosciences, 35(2), 231–240. https://doi.org/10.1007/S12038-010-0027-1,
Rao, A. S., Rajanikanth, B., & Seshadri, R. (1989). Volatile Aroma Components of Curcuma amada Roxb. Journal of Agricultural and Food Chemistry, 37(3), 740–743. https://doi.org/10.1021/JF00087A036/ASSET/JF00087A036.FP.PNG_V03
Reimand, J., Isserlin, R., Voisin, V., Kucera, M., Tannus-Lopes, C., Rostamianfar, A., Wadi, L., Meyer, M., Wong, J., Xu, C., Merico, D., & Bader, G. D. (2019). Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols, 14(2), 482–517. https://doi.org/10.1038/S41596-018-0103-9,
Ren, W., Wulan, T., Dai, X., Zhang, Y., Jia, M., Feng, M., & Shi, X. (2024). Study on the interaction between volatile oil components and skin lipids based on molecular docking techniques. Digital Chinese Medicine, 7(2), 148–159. https://doi.org/10.1016/J.DCMED.2024.09.006
Rout, T., Mohapatra, A., Kar, M., Patra, S., & Muduly, D. (2024). Centrality Measures and Their Applications in Network Analysis: Unveiling Important Elements and Their Impact. Procedia Computer Science, 235, 2756–2765. https://doi.org/10.1016/J.PROCS.2024.04.260
Sabaawy, H. E., Ryan, B. M., Khiabanian, H., & Pine, S. R. (2021). JAK/STAT of all trades: linking inflammation with cancer development, tumor progression and therapy resistance. Carcinogenesis, 42(12), 1411–1419. https://doi.org/10.1093/CARCIN/BGAB075
Sahoo, S., Jyotirmayee, B., Nayak, S., Samal, H. B., & Mahalik, G. (2023). Review on Phytopharmacological Activity of Curcuma amada Roxb. (Mango ginger). In Defence Life Science Journal (Vol. 8, Issue 4, pp. 351–362). Defense Scientific Information and Documentation Centre. https://doi.org/10.14429/dlsj.8.18793
Sever, R., & Brugge, J. S. (2015). Signal Transduction in Cancer. Cold Spring Harbor Perspectives in Medicine, 5(4), a006098. https://doi.org/10.1101/CSHPERSPECT.A006098
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research, 13(11), 2498–2504. https://doi.org/10.1101/gr.1239303
Srivastava, A. K., Srivastava, S. K., & Shah, N. C. (2001). Constituents of the Rhizome Essential Oil of Curcuma amada Roxb. from India. Journal of Essential Oil Research, 13(1), 63–64. https://doi.org/10.1080/10412905.2001.9699608;WGROUP:STRING:PUBLICATION
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., Gable, A. L., Fang, T., Doncheva, N. T., Pyysalo, S., Bork, P., Jensen, L. J., & Von Mering, C. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Research, 51(1 D), D638–D646. https://doi.org/10.1093/nar/gkac1000
The PyMOL Molecular Graphics System. (n.d.). PyMOL | pymol.org. Retrieved November 7, 2023, from https://pymol.org/2/
Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. https://doi.org/10.1002/jcc.21334
Valls, P. O., & Esposito, A. (2022). Signalling dynamics, cell decisions, and homeostatic control in health and disease. Current Opinion in Cell Biology, 75, None. https://doi.org/10.1016/J.CEB.2022.01.011
Wang, Y. L., Liu, H. F., Shi, X. J., & Wang, Y. (2018). Antiproliferative activity of Farnesol in HeLa cervical cancer cells is mediated via apoptosis induction, loss of mitochondrial membrane potential (ΛΨm) and PI3K/Akt signalling pathway. Journal of B.U.ON., 23(3), 752–757. https://www.researchgate.net/publication/326399147_Antiproliferative_activity_of_Farnesol_in_HeLa_cervical_cancer_cells_is_mediated_via_apoptosis_induction_loss_of_mitochondrial_membrane_potential_LPsm_and_PI3KAkt_signalling_pathway
Xue, C., Yao, Q., Gu, X., Shi, Q., Yuan, X., Chu, Q., Bao, Z., Lu, J., & Li, L. (2023). Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer. Signal Transduction and Targeted Therapy 2023 8:1, 8(1), 1–24. https://doi.org/10.1038/s41392-023-01468-7
Zambuzzi, W. F., Coelho, P. G., Alves, G. G., & Granjeiro, J. M. (2011). Intracellular signal transduction as a factor in the development of “Smart” biomaterials for bone tissue engineering. Biotechnology and Bioengineering, 108(6), 1246–1250. https://doi.org/10.1002/BIT.23117;WGROUP:STRING:PUBLICATION
Ziaei, A., Ramezani, M., Wright, L., Paetz, C., Schneider, B., & Amirghofran, Z. (2011). Identification of spathulenol in Salvia mirzayanii and the immunomodulatory effects. Phytotherapy Research, 25(4), 557–562. https://doi.org/10.1002/PTR.3289
Authors

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. Copyright @2017. This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (http://creativecommons.org/licenses/by-nc-sa/4.0/) which permits unrestricted non-commercial used, distribution and reproduction in any medium